
Reserch
Institute for Molecular and Cellular Regulation
Laboratory of Genome Science

Izuho Hatada Professor
Epigenetics
Research and Education
1) Epigenome analysis of diseases

It has been long time after starting extensive genetic analysis of human diseases. However, some of the diseases are found not to be caused by genetic changes rather by the alteration of epigenome which is the switch of the genes. Aberrant changes of epigenome 驍オ・イ・つcaused by environments results in several diseases like diabetes. We are going to clarify the causative genes for diabetes, obese and cancer by massive analysis of the epigenome (Fig. 1). We identified several candidate obese genes, which epigenetically regulate the differentiation of adipose cells including WGEF. We have been analyzing the function of these genes by making knockout mice by high efficienct CRISPR/Cas method.
2) Development of tumor-free pluripotent stem cells – New choice of regenerative medicine
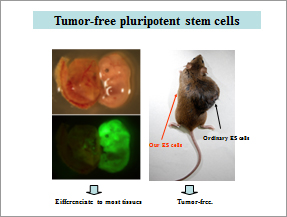
Differentiated tissues derived from pluripotent stem cells (ES & iPS cells) unfortunately form tumors with considerable frequency after transplantation therapy. In most cases, oncogenes are epigenetically activated in these tumors. We established new pluripotent stem cells that are resistant to epigenetic activation of oncogenes. These cells do not form tumors after transplantation (Fig. 2). We have been trying to making tumor-free iPS cells by disruption of these oncogenes by high efficienct CRISPR/Cas method.
3) Epigenetic anomalies in assisted reproductive technology and regenerative medicine
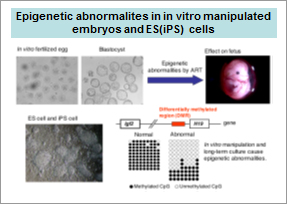
A lot of couples have children by the favor of assisted reproductive technology (ART); however, epigenetic anomalies are often seen in these in vitro manipulated embryos (Fig. 3). Methylation anomalies increase the risk of diseases such as Beckwith-Wiedemann Syndrome (BWS). ES cells and iPS cells cultured for a long term in vitro also show methylation anomalies. We are now studying why these anomalies occurred in vitro.
Recent Publication
- Horii T, Tamura D, Morita S, Kimura M, Hatada I: Generation of an ICF Syndrome Model by Efficient Genome Editing of Human Induced Pluripotent Stem Cells Using the CRISPR System. Int. J. Mol. Sci. 14:19774-19781 (2013)
- Morita S, Horii T, Kimura M, Hatada I: MiR-184 regulates insulin secretion through repression of Slc25a22. PeerJ 1: e162 (2013)
- Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I: miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci.,14: 14647-14658 (2013)
- Takahashi M, Kamei Y, Ehara T, Yuan X, Suganami T, Takai-Igarashi T, Hatada I, Ogawa Y: Analysis of DNA methylation change induced by Dnmt3b in mouse hepatocytes. Biochem Biophys Res Commun. 434:873-878 2013 (2013)
- Gailhouste L, Gomez-Santos L, Hagiwara K, Hatada I, Kitagawa N, Kawaharada K, Thirion M, Kosaka N, Takahashi RU, Shibata T, Miyajima A, Ochiya T: MiR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 58:1153-1165 (2013)
- Deguchi K, Nagamatsu G, Miyachi H, Kato Y, Morita S, Kimura H, Kitano S, Hatada I, Saga Y, Tachibana M, Shinkai Y: Posttranscriptional Regulation of Histone Lysine Methyltransferase GLP in Embryonic Male Mouse Germ Cells. Biol Reprod 88:3 (2013)
- Morita S, Takahashi RU, Yamashita R, Toyoda A, Horii T, Kimura M, Fujiyama A, Nakai K, Tajima S, Matoba R, Ochiya T, Hatada I: Genome-Wide Analysis of DNA Methylation and Expression of MicroRNAs in Breast Cancer Cells. Int J Mol Sci. 13:8259-8272 (2012)
- Kobayashi Y, Aizawa A, Yoshizawa C, Gotoh T, Horiguchi H, Souma H, Ikeuchi Y, Kakegawa S, Watanabe T, Maruyama K,Takizawa T, Morikawa A, Hatada I, Arakawa H: DNA Methylation Changes between Relapse and Remission of Minimal Change Nephrotic Syndrome. Pediatr Nephrol. 27:2233-2241 (2012)
- Sato N, Yamakawa N, Masuda M, Sudo K, Hatada I, Muramatsu M.: Genome-wide DNA methylation analysis reveals phytoestrogen modification of promoter methylation patterns during embryonic stem cell differentiation. PLoS One 6: e19278 (2011)
- Horii T, Hatada I. Reprogrammed parthenogenetic ES cells – new choice for regenerative medicine. Methodological Advances in the Culture, Manipulation and Utilization of Embryonic Stem Cells for Basic and Practical Applications, INTECH, 221-236 (2011)
- Horii T, Suetake I, Yanagisawa E, Kimura M, Morita S, Nagao Y, Imai H, Tajima S, Hatada I: The Dnmt3b splice variant is specifically expressed in in vitro-manipulated blastocysts and their derivative ES cells. J Reprod Dev 57:579-585 (2011)
- Obata Y, Hiura H, Fukuda A, Komiyama J, Hatada I, Kono .: Epigenetically Immature Oocytes Lead to Loss of Imprinting During Embryogenesis. J Reprod Dev 57: 327-334 (2011)
- Horii T, Yanagisawa E, Kimura M, Morita S, Hatada I: Epigenetic differences between embryonic stem cells generated from blastocysts developed in vitro and in vivo. Cell Reprogram. 12:551-563 (2010)
- Horii T, Morita S, Kimura M, Hatada I: Epigenetic regulation of adipocyte differentiation by a Rho guanine nucleotide exchange factor, WGEF. PLoS One. 4:e5809 (2009)
- Morita S, Hara A, Kojima I, Horii T, Kimura M, Kitamura T, Ochiya T, Nakanishi K, Matoba R, Matsubara K, Hatada I: Dicer is required for maintaining adult pancreas. PLoS One. 4:e4212 (2009)
